I am happy to report that OnScreen DNA Replication, my iPad app that simulates the process in its title is now available for purchase and download. In joining the previously released OnScreen DNA Model and OnScreen Gene Transcription on the App Store, it completes the suite of interactive apps designed to teach the details of DNA’s structure and function using the same three-dimensional model.
The guiding concept of these OnScreen DNA apps is that seeing the basic molecular components of nucleic acids in a sufficiently detailed three-dimensional ball-and-stick model, one that requires unwinding the strands of DNA before they can used as templates for daughter strand or messenger RNA construction by complementary base pairing (also shown, of course), will foster an intuitive grasp of Nature’s beautiful solution to the problems of critical biological information storage, retrieval, and inheritance. The necessity of enzymes for the processes is also emphasized in a conspicuous way.
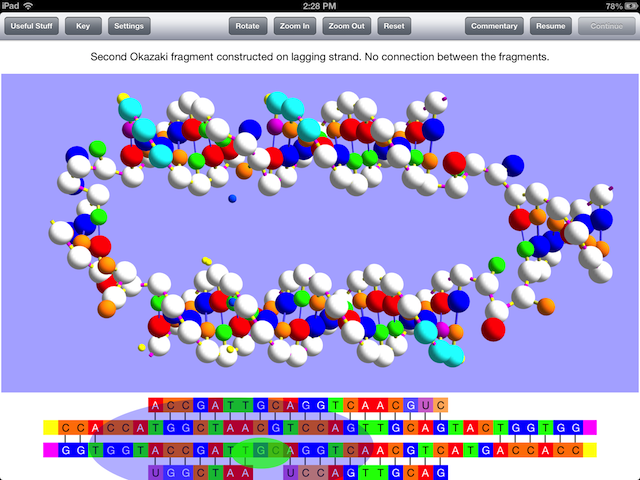
There is a thirty-second video excerpt of the simulation online that shows the part of the simulation seen in the image above, which should give an idea of how the model is used to display the processes that occur in replication, but note that the video was made with an iPad simulator and runs more slowly than the actual app on an iPad does. Unfortunately, Firefox can’t show it.
Just to summarize the OnScreen DNA Replication iPad app’s key features and advantages, I note that it:
- Shows three-dimensional, color-coded double helix structures, not two-dimensional ladders.
- Uses animations that show hydrogen bonds being formed (as sticks connecting base pairs in the ball-and-stick representation) and broken.
-
Models proper right-handed DNA (depressing how many images and even simulations depict left-handed DNA).
-
Indicates the enzymes enabling the reactions shown and where they are acting.
- Includes background material in a popup view.
- Shows every major step in replication, with commentary available in a popup view.
- Provides the option to run the simulation without pause (except when the user intervenes) or to automatically pause after key steps in order to conveniently read commentary if desired.
- Provides a key to the color code etc. in a popup view.
- Provides a visual representation of strand polarity.
- Maps the nucleotide-base sequence of the 3D model to a GCAT base-by-letter linear representation.
- Deals with the end problem: telomerase reverse transcription shown.
- Calls attention to the crucial action of the enzyme pyrophosphatase.
- Allows the user to zoom in or out and rotate the model by touchscreen gestures to see it from different perspectives even as the simulation is running.
- Is suitable for just about anyone wanting to learn how DNA works, from middle school students to intellectually curious adults, since no advanced chemistry knowledge is assumed.
The replisome enzymes responsible for replication are identified, but their visual representation is confined to the linear (GCAT) sequence view. This has the advantage of making the point that the process requires the enzyme, while showing the location of its catalytic activity, but without obscuring the basic structural changes that are occurring in the model view. The RNA of the enzyme telomerase, however, is shown in the model view as well as in the sequence view, since there is base-pairing to be seen in the model view during the reverse transcription process.
Let me mention some features of DNA replication that can be difficult to grasp, which I think the app’s simulations convey clearly. Nature has not provided the cell with an enzyme for beginning the construction of a daughter DNA strand with a DNA nucleotide. A DNA nucleotide when it is paired to a nucleotide in the template strand must also be connected at its phosphate-bearing end to a nucleotide already present in the daughter strand. There is an enzyme to begin a daughter strand with RNA nucleotides, however, and this is utilized in DNA replication. The construction of “primer RNA” is of course shown in the simulations of OnScreen DNA Replication. RNA’s point of difference from DNA, the different sugar-phosphate backbone, stands out by virtue of its color in the model. The simulation shows three RNA nucleotides in each primer RNA chain, which is smaller than the number in Nature, but long enough to illustrate the principle. The app is a teaching model, not a perfect mapping of reality in every detail.
This requirement of primer RNA is why the so-called lagging daughter strand of DNA is constructed with a series of Okazaki fragments from which the RNA must be replaced by DNA and a final connection made between the fragments. If you don’t know what leading and lagging strand refers to or what Okazaki fragments are, OnScreen DNA Replication will teach you, while showing all the steps and enzymes required in their construction and modification.
The necessity for starting a new Okazaki fragment with primer RNA leads to the “end problem” in the replication of linear (not circular) DNA. I encountered this problem for the first time in a very practical way when I was programming simulations for OnScreen DNA for the Macintosh a few years ago. Okazaki fragments could be dealt with by having the primer RNA replaced by DNA–except at the very end of the lagging strand. What happened there? I had to do a good bit of digging to find out how to deal with the problem, since introductory treatments of DNA replication ignored it altogether. I learned then how the enzyme telomerase solved the problem, so I added telomerase’s action to the simulation. Since telomerase makes use of reverse transcription to extend the lagging DNA strand, the demonstration of that process, which is also utilized by retroviruses, is a bonus. Telomeres and how telomerase prevents strand-shortening are discussed both in the Useful Stuff and in the Commentary popup views.
OnScreen DNA Replication also shows a crucial step in nucleic acid polymerization that is usually ignored in introductory treatments: the reaction that breaks into two phosphate molecules the pyroposphate molecule which is a by-product of the polymerization. WIthout this splitting of the pyrophosphate molecule into the two phosphates, which is brought about by the action of the pyrophosphatase enzyme, the reaction to reverse the polymerization (that reverse reaction being thermodynamically favored to occur) would make life that utilizes chains of nucleic acids impossible. Since the two-phosphate state is even more highly favored over pyrophosphate, catalyzing the splitting of pyrophosphate makes the overall chain of reactions practically irreversible. Pyrophosphatase also performs this life-saving action for other reactions in the cell, but this is the one of immediate concern in DNA replication.
We highlight this crucial step by representing in the model view the pyrophoshate given off with every formation of a new phosphodiester bond as a newly appearing ball traveling away from the reaction site and then splitting into two smaller balls. This is meant both to arouse the curiosity of the observer to read about what is happening and to reinforce the necessity of this step. A water molecule is also shown leaving the site of polymerization just to plant the idea that a condensation reaction has occurred, as is the case in the synthesis of all the important biological macromolecules. You can see this in the video clip linked to above. The water molecule and pyrohphosphate breakup are also shown in the OnScreen Gene Transcription iPad app’s simulation of messenger RNA construction.
The description of OnScreen DNA Replication as it appears on the app store follows.
Looks great on an iPad Mini as well as “full-size” iPads. Go see the short video excerpt on the nondummies.com website. We know of no other simulation, app or internet, that shows what happens in DNA replication as thoroughly as this app does. OnScreen DNA Replication shows all of the several steps (indicating the corresponding enzymes responsible for those steps) necessary for one double helix to become two identical to the original. Through the use of engaging 3D animations with a virtual double helix model (not a 2D ladder) it makes clear and memorable how DNA daughter strands are constructed nucleotide by nucleotide in replication.
Students from middle school on up can learn from the app, as no advanced knowledge of chemistry is assumed. The model is exactly the same as the one found in OnScreen DNA Model, a companion app that teaches the structural details of DNA, and in OnScreen Gene transcription, another companion app that shows how protein recipes are copied into messenger RNA. Detailed commentary on what the animations demonstrate in each step is available in a popover view, and a wealth of background material is to be found in a “Useful Stuff” popover.
The sequence of events in DNA replication unfold in three-dimensional simulations that don’t skip over the need for unwinding the DNA after the strands have been separated. The formation of a hybrid DNA-RNA double helix during the first step of primer RNA construction is correctly shown. DNA and RNA nucleotides are seen to move into place and then form hydrogen bonds with their base-pair mates in the template DNA strand. Important details about replication that are often given short shrift or omitted altogether, such as the essential role the enzyme pyrophosphatase plays in the cell, are included.
The concepts of leading and lagging strands and what the Okazaki fragments are and how they are constructed and then joined together through the actions of various replisome enzymes are made clear and memorable through the three-dimensional simulations in the Model View and the representations of enzymes in the Sequence View.
The “end problem” of linear DNA strand replication is not swept under the rug as often happens. Instead, the basic principle of how the enzyme telomerase uses its own RNA to extend the lagging DNA strand by means of reverse transcription is illustrated using simple models with only the RNA and DNA showing.
Set the simulation to pause after each new significant step or pause it only when you want. Commentary on what is happening is literally at your fingertip in a popover. Rotate, translate, or zoom the model during the simulated replication for a better view just by finger slide gestures.
The ball-and-stick model has the advantage of clarity at the expense of atomic detail. The replisome enzyme complex, while not shown in the view with the DNA model, so as not to obscure what is happening with bonds and strands, is depicted in the Sequence View below the model, thus making the point that it moves along the DNA, as it initiates and controls the reactions in replication. Furthermore, the actions of the individual enzymes that make up the replisome are also indicated in the Sequence View.
For efficient and enjoyable learning about DNA’s structure and how it works both to pass on protein recipes in transcription (messenger RNA construction) and to replicate itself into two double helix structures identical to the original, I confidently recommend the three apps OnScreen DNA Model, OnScreen Gene Transcription, and now OnScreen DNA Replication.


 OnScreen
OnScreen
 OnScreen
OnScreen OnScreen
OnScreen